Abstract
Genetic aberrations in leukemic blasts are associated with clinical outcomes in patients with AML. However, the role of inherited genetic variants with regards to variation in AML treatment outcome remains obscure. To that end, we conducted a genome wide association study (GWAS) on 400 newly diagnosed pediatric AML patients to identify single nucleotide polymorphism (SNPs) that may be associated with minimal residual disease (MRD1) after induction 1 chemotherapy, event-free survival (EFS) and overall survival (OS).
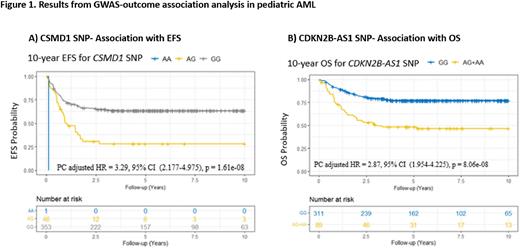
In this study, we utilized whole-genome genotyping with Illumina's Omni2.5Exome-8 BeadChip array to investigate SNPs for association with clinical outcome in 2 pediatric AML cohorts: the multi-site AML02 [NCT00136084, n=167] and AML08 [NCT00703820, n=233] trials. Post GWAS quality control, 1,365,439 unphased SNP genotypes were selected for imputation using TOPMed Imputation Server. Following imputation and additional quality control filtering, 6,741,200 SNPs evaluated for association with clinical outcomes (MRD1, EFS and OS). To improve statistical rigor, two GWAS approaches were utilized: i) A one-stage GWAS with both clinical trial cohorts combined (n=400) and ii) A two-stage GWAS where we utilized a two-step iterative resampling procedure (PMID: 26377241) by splitting the cohort at a 70:30 ratio into discovery and replication cohorts, balanced by clinical trial, treatment arm and initial risk group at diagnosis (100 randomizations performed). For each of the 100 iterations of the two-stage GWAS, we used the discovery cohort to perform a genome-wide analysis, followed by a replication screen. SNPs replicated in at least 20 of 100 rounds of discovery-replication were prioritized. A comparison of the two approaches showed all significant SNPs in the two-stage GWAS were also captured in the one-stage GWAS. For EFS, a Bonferroni-adjusted genome-wide significant association was observed for a SNP in CSMD1 and this SNP was replicated in 83 out of 100 discovery-replication rounds for two-stage GWAS. For CSMD1 SNP, presence of variant A allele has significantly reduced EFS (HR=3.29, 95% CI (2.177-4.975), P=1.61x10-8, Figure 1A). CSMD1 is a membrane-bound complement inhibitor that has been shown to act as a putative tumor suppressor gene in other malignancies (PMIDs:27764775, 35623420). Another significant marker worth mentioning was an intronic SNP in CDKN2B-AS1 that associated with poor outcome [EFS: HR=2.37, 95% CI (1.700-3.317), P=4.40x10-7] and [OS: HR=2.87, 95% CI (1.954-4.225), P=8.06x10-8, Figure 1B]. This marker is in high linkage disequilibrium with 26 other SNPs within the CDKN2B and CDKN2B-AS1 cluster. The product of this gene is an RNA molecule that interacts with polycomb repressive complex-1 (PRC1) and -2 (PRC2) involved in silencing other genes within this cluster through epigenetic mechanisms (PMID: 20716771). CDKN2B-AS1 has been reported to play a role in tumorigenesis and chemoresistance in pediatric T-cell acute lymphoblastic leukemia and with the risk of chronic lymphocytic leukemia (PMIDs: 32976214/23770605). Though limited by numbers, this is one of the largest GWAS conducted in pediatric AML to identify SNPs associated with clinical outcomes. Our top results show that genetic variation in several genes of biological relevance may impact MRD1, EFS and OS. Future studies are aimed at looking at SNP/gene and association with gene expression collected from leukemic cells at time of diagnosis to establish AML-specific eQTLs and functional validation of the most promising candidates. Additionally, future analyses will include other subgroups such as cytogenetic risk group and validation of these findings in larger cohort of patients.
Disclosures
Inaba:Servier: Other: Grants and Personal Fees; Amgen: Other: Grants and Personal Fees; Incyte: Other: Grants; JAZZ: Other: Personal Fees; Chugai: Other: Personal Fees. Pui:Adaptive Biotechnologies: Membership on an entity's Board of Directors or advisory committees; Novartis: Other: Data monitoring committee.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal